Table of Contents
If you want to generate visualizations for your experiments (see How to conduct experiments with GCG), you should use the automated generation features of our Visualization Suite. However, you might need to generate single visualizations manually, which is what is explained on this page.
Visualization Generation
Many different things can be plotted using our visualization framework. You can find all possible arguments here.
Common Prerequisites
In this section, we describe what is required to generate the visualizations using the included visualization scripts. They are also required to be fulfilled if you are only using the Visualization Suite.
Software requirements
The scripts will mostly give helpful error messages if you have misconfigured anything, so please pay attention to their output.
Python
Except for the comparison_table script, all visualizations are generated using python. For those, you need python3 to be installed correctly on your system:
sudo apt-get install python3
Python-Packages
For each script, you will need packages and some are probably not yet installed on your computer. You can do that by first installing pip:
sudo apt-get install python3-pip
And then the needed packages via
pip3 install numpy matplotlib pandas sklearn
Furthermore, we recommend to install the package
tqdm
which shows progress bars which can be useful in particular for very large runtime data.
LaTeX
For the pricing visualizations, a working LaTeX installation is required, i.e.
sudo apt-get install dvipng texlive-latex-extra texlive-fonts-recommended
should be up to date.
Run tests
First, you have to do a testrun to gather the data for the visualizations. A guide on how to do that can be found here.
The scripts sometimes expect a certain flag to be set. The following table gives an overview over the requirements for the runtime data for each visualization script.
| Expected Input | Required compile flags | Required test flags | |
|---|---|---|---|
| Performance Profile | .out/.res | - | - |
| General | .out | - | STATISTICS=true |
| Bounds | .out | STATISTICS=true | STATISTICS=true |
| Classifier/ Detection | .out | - | DETECTIONSTATISTICS=true |
| Pricing | .out, .vbc | STATISTICS=true | - |
| Tree | .vbc | - | |
| Comparison Table | .res | - | - |
Generate Visualizations Manually
You can create different plots with the included scripts in the
statsfolder. All scripts are briefly explained in the following. When describing how to execute them, it is always assumed that you are inside thestatsfolder.
Common arguments
The following arguments are common across all following scripts (except for the performance profile plotter and comparison table).
Defining the Output Directory
-o OUTDIR, --outdir OUTDIR
output directory (default: "plots")
Parsing without Plotting, Plotting without Parsing
-save, --savepickle parses the given .out-file without plotting -load, --loadpickle loads the given .pkl-file and plots it
Performance Profile Plotter
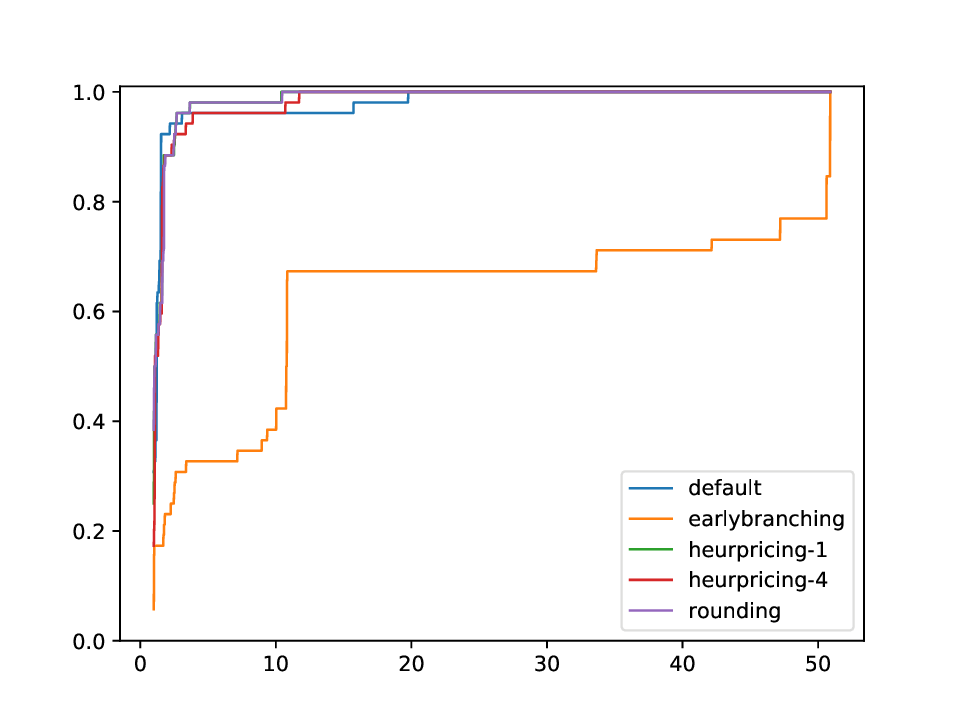
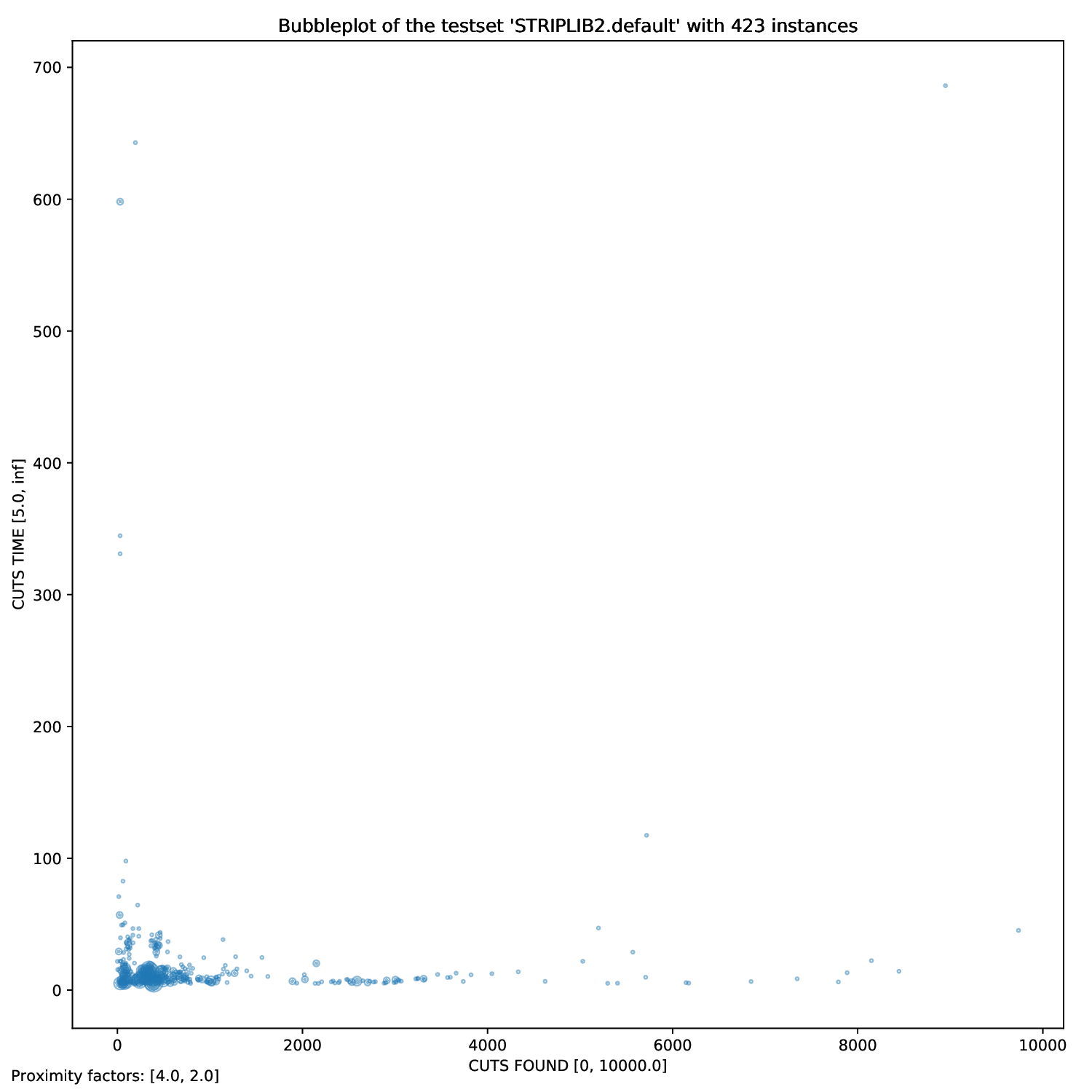
Using this plotter, one can generate performance profiles. In those, the x-axis represents the factor by which the respective run is worse than the optimal run, while the y-axis is the corresponding probability.
Execution
python3 general/performance_profile.py FILES
with FILES being some (space-separated) .res or .out files in the format as shown in 'What files do I get'.
General Plotter
The general plotter is able to plot two or more arguments parsed by the general_parser.py script in different ways. You can find all parsed arguments here.
Execution
python3 general/{bubble,plot,twin,time}.py [args] FILES
with FILES being either some pickles, or some outfiles, and [args] being as defined below.
Arguments
Defining Data to be plotted
Times are the most common arguments to plot (that's the reason for the naming), but you can use whatever argument parsed you want. You can find a list of those here.
You can define what data to plot with the --times argument:
-t [TIMES [TIMES ...]], --times [TIMES [TIMES ...]]
times to be plotted.
Whereas
- for
bubble.pyandtwin.py, you may define exactly two times (or none),
- for
plot.pyyou can define as many times as you like and - for
time.pyyou can also define an arbitrary amount, but the "OTHERS" time will be added automatically.
Defining Filters for the Data
For all arguments, you can define filters. For example, if you only want to plot all those instances that had a Detection Time of between 10 and 20 seconds, you simply add those bounds to the time argument:
python3 general/time.py check.gcg.out -A -t "DETECTION TIME" 10 20 "RMP LP TIME"
In this example, we generate all plots (-A) for all instances that had a detection time between 10 and 20 seconds in the check.gcg.out file with the arguments "DETECTION TIME" and "RMP LP TIME" (without filter).
Defining additional Arguments (only for time.py)
For the time.py script, you can define which plots you want to generate. The arguments are as follows:
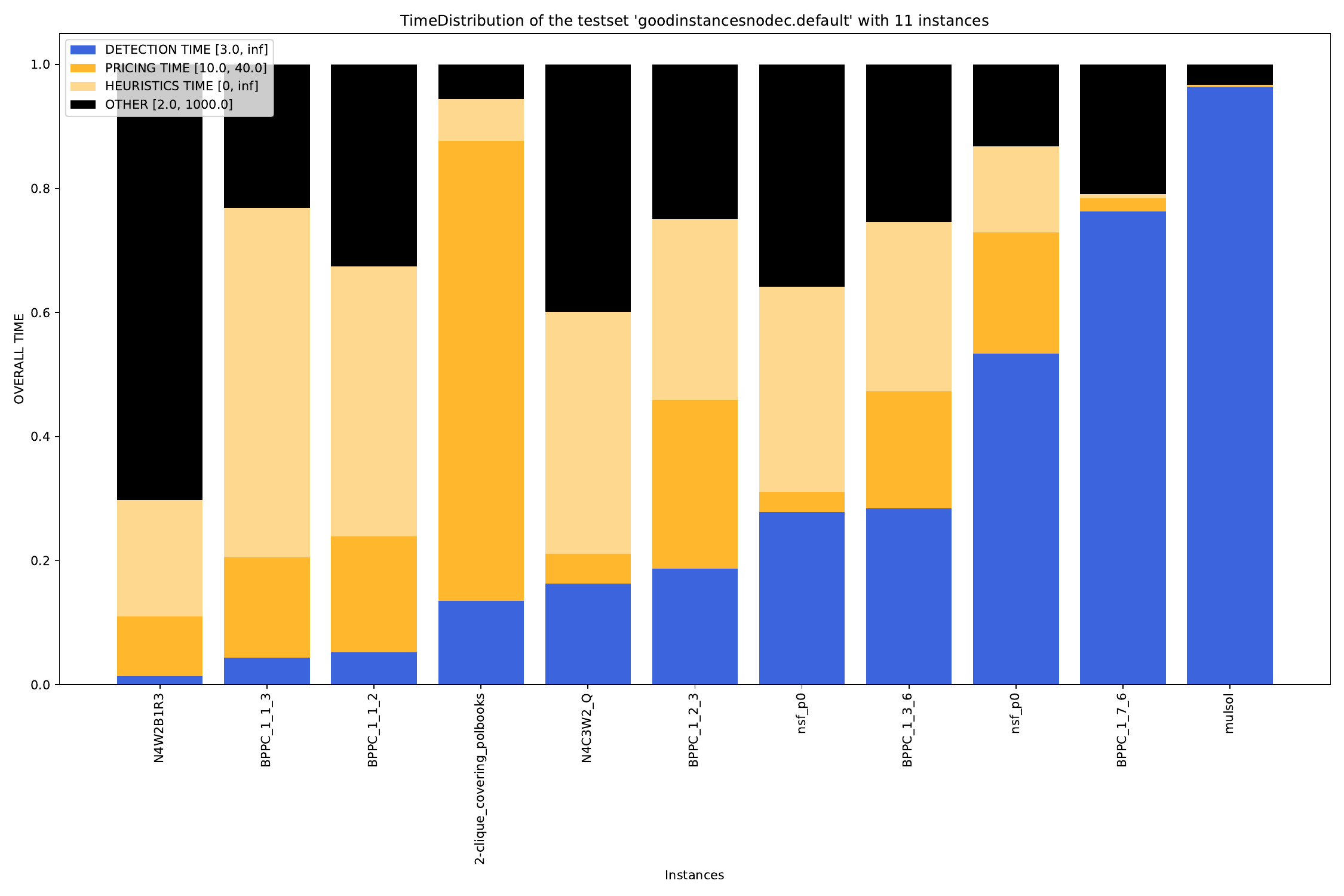
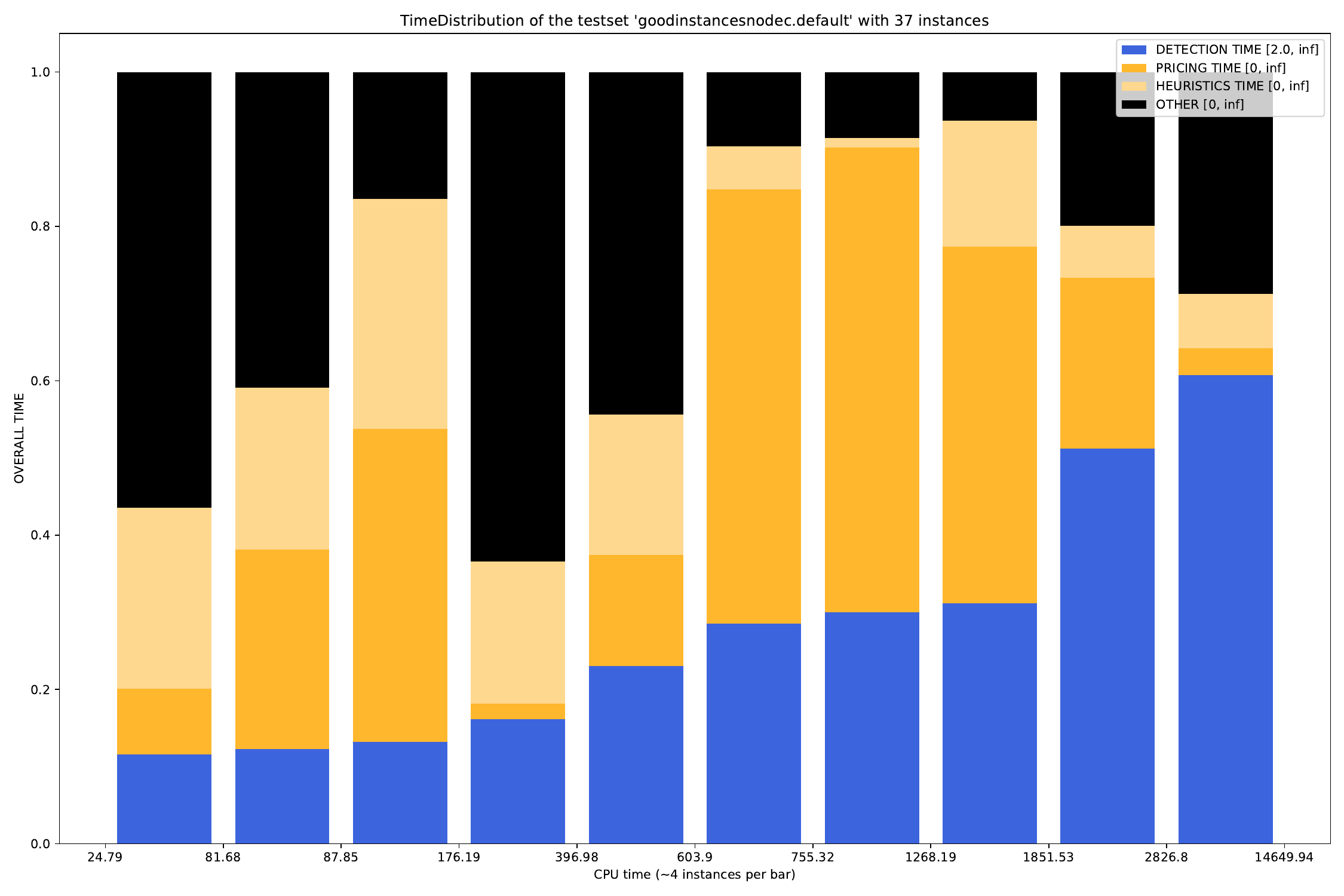
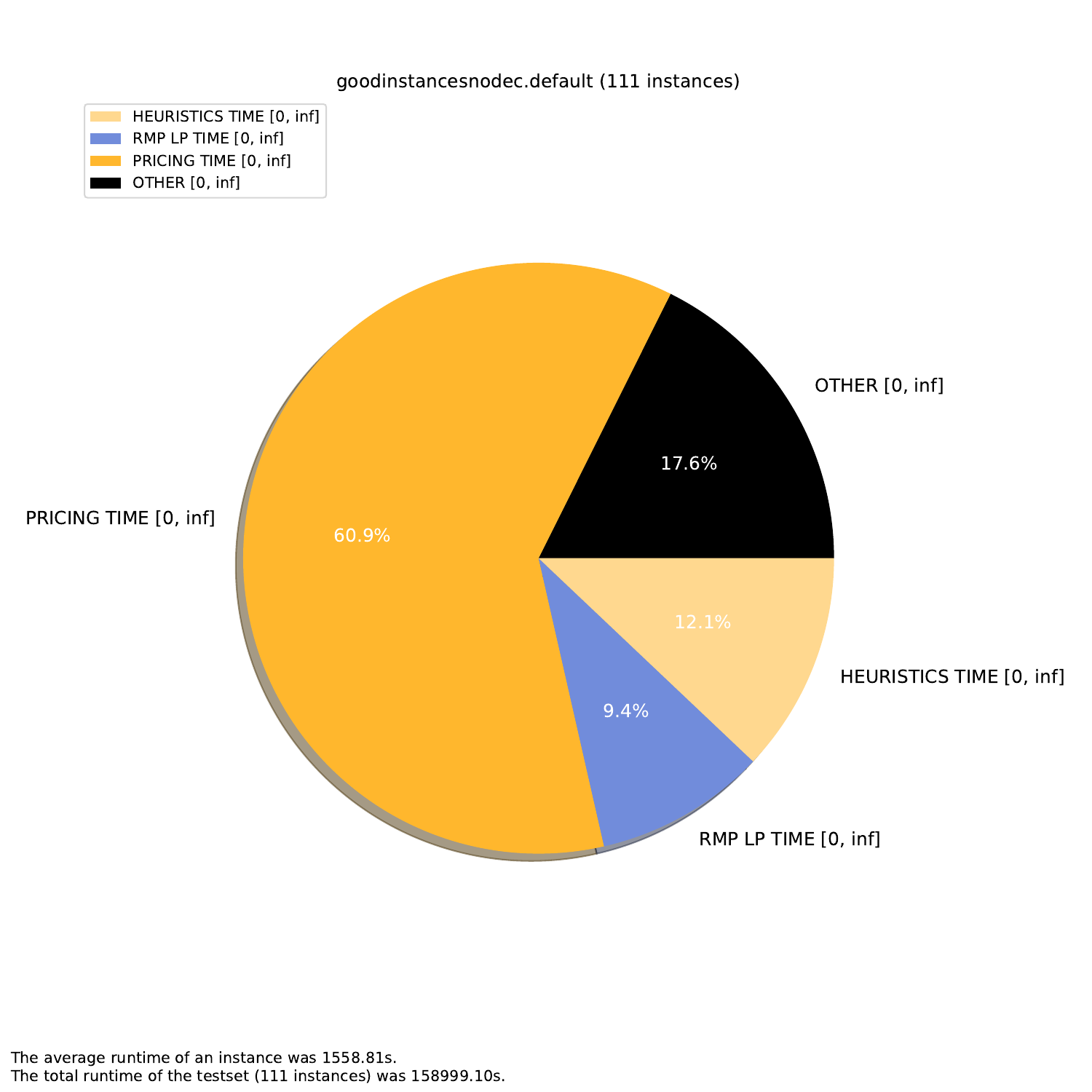
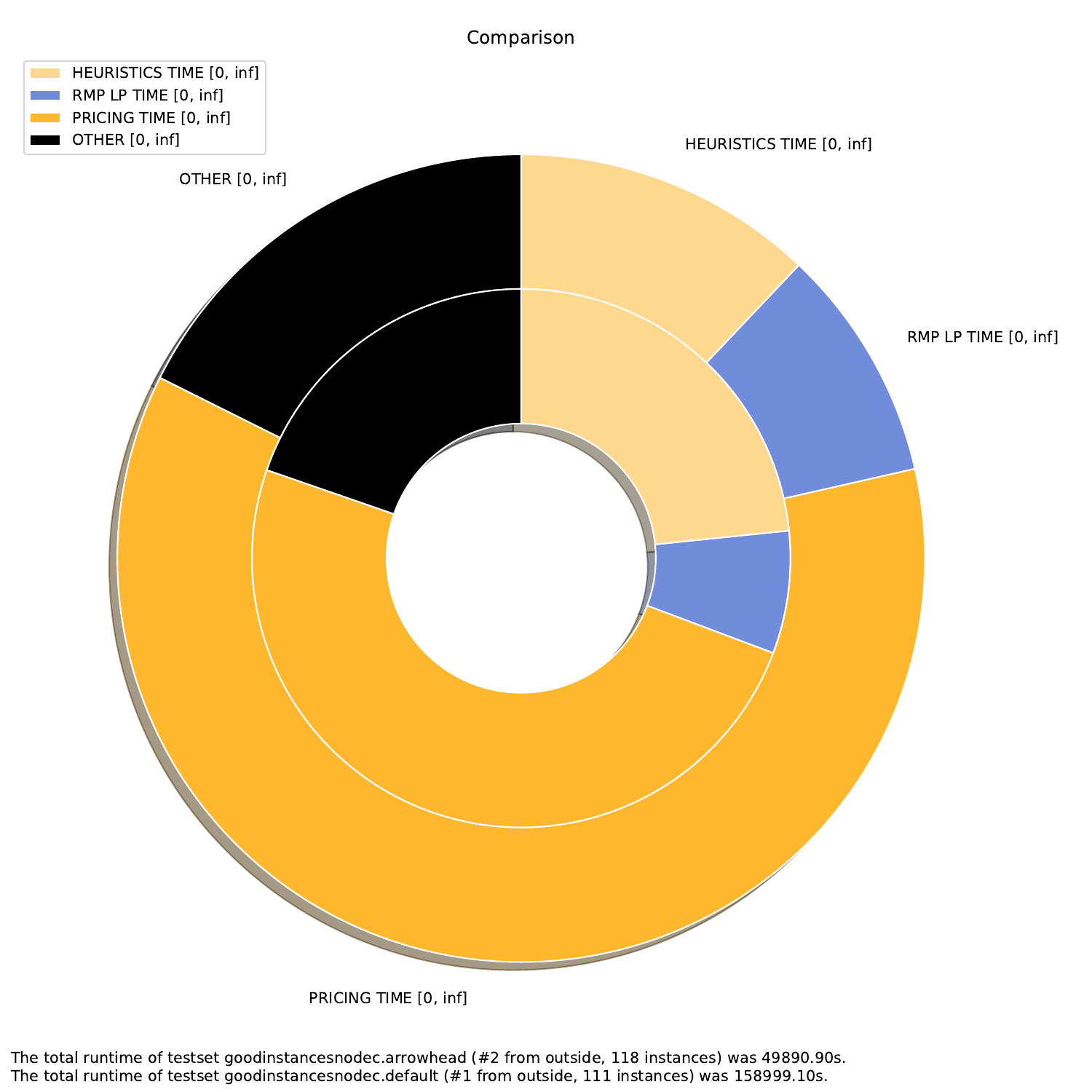
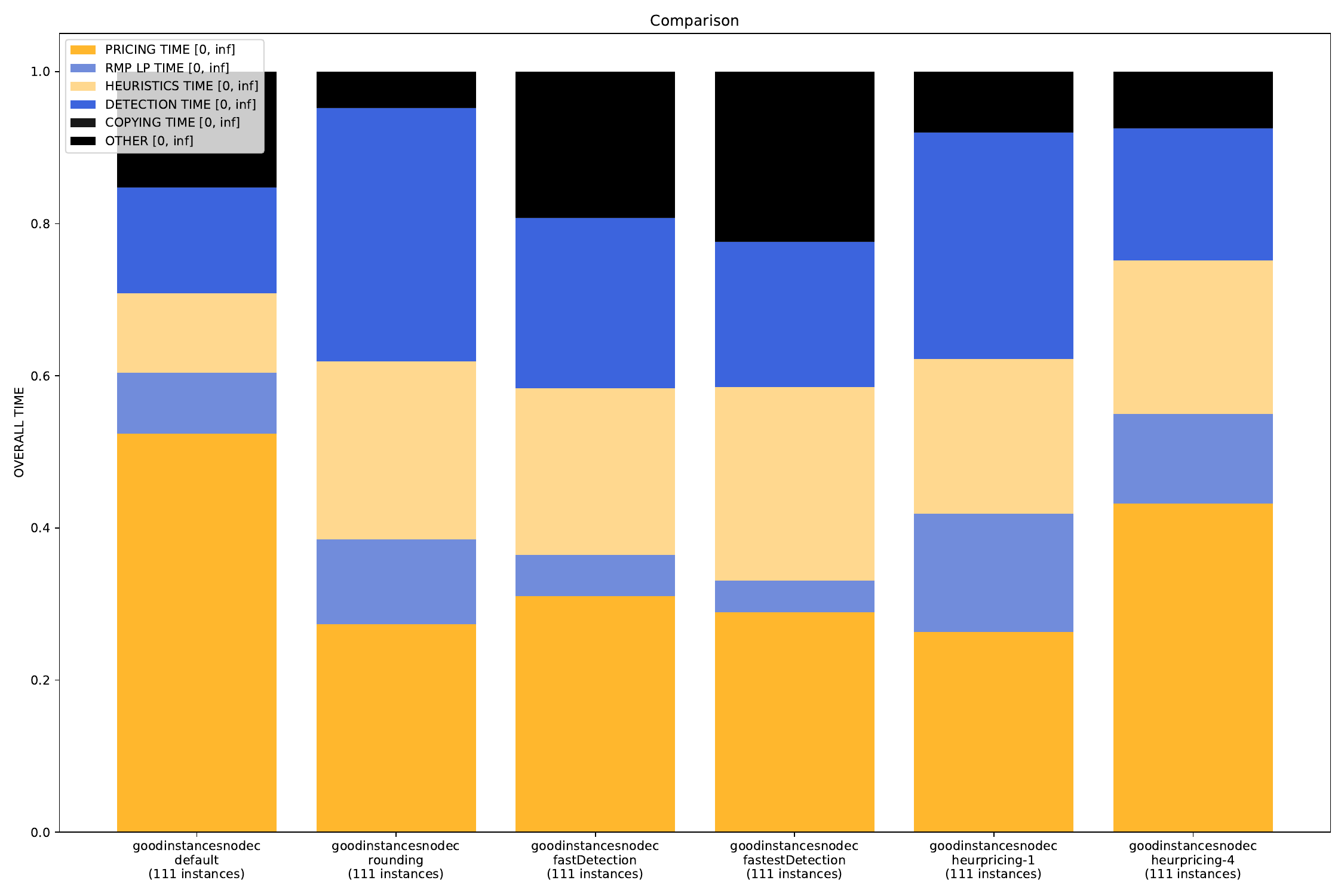
-A, --all create all visualizations -B, --bar create barchart -G, --grouped-bar create barchart, grouped by CPU times -P, --plot create simple plot --pie create a pie chart plot (will automatically activate --single (see below))
Additionally, for the grouped-bar plot, you may define a number of buckets, in which the script will automatically sort the (nearly) same number of instances into.
--buckets BUCKETS amount of buckets (resolution of grouped_bar plot)
Finally, you can compare two different runs. Though recommended, it is not required for the runs to have been on the same testset.
--single if set, all outfiles will be summed and cumulated in a single plot --compare if set, each outfile will be summed and plotted with all other outfiles
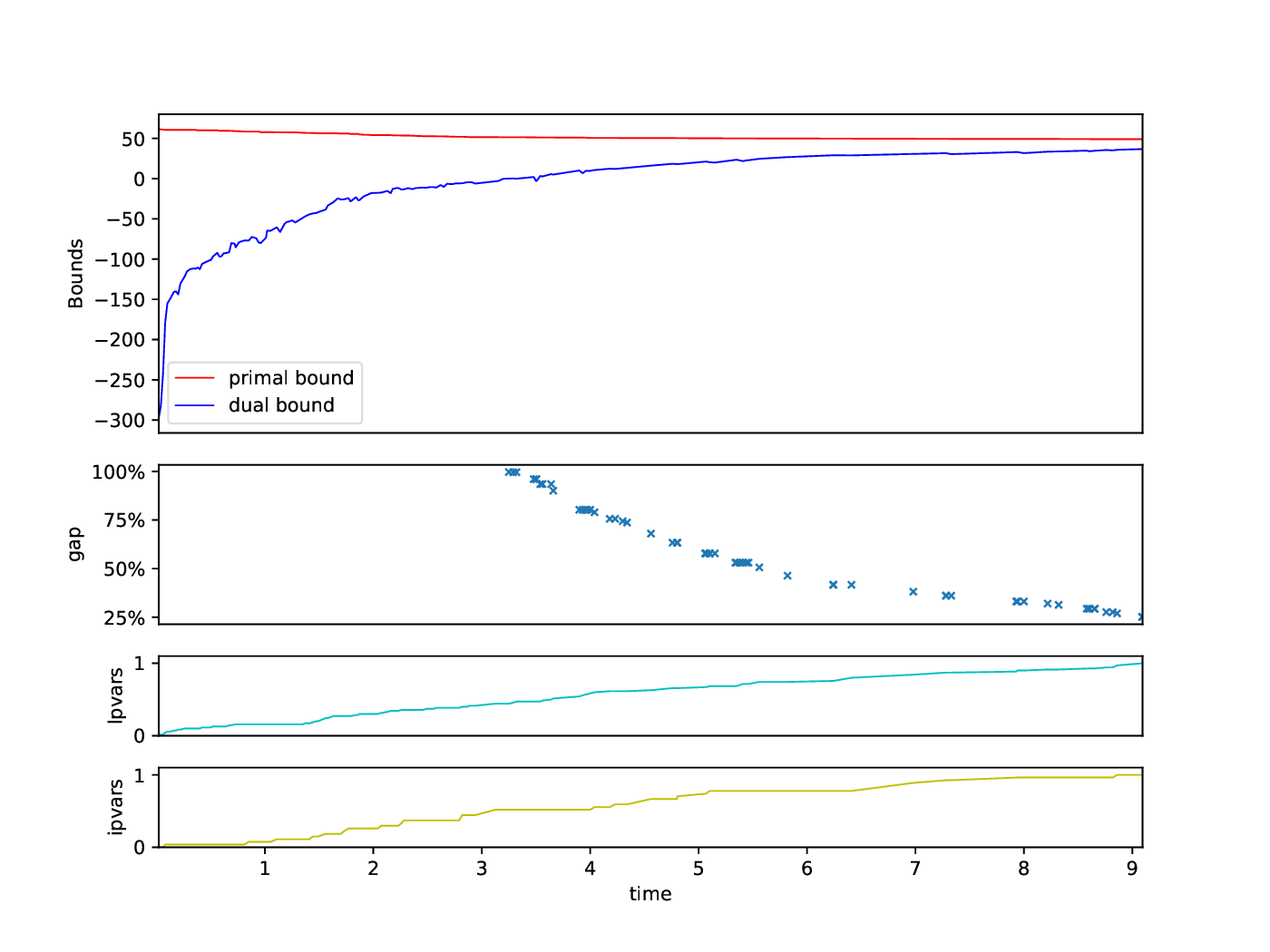
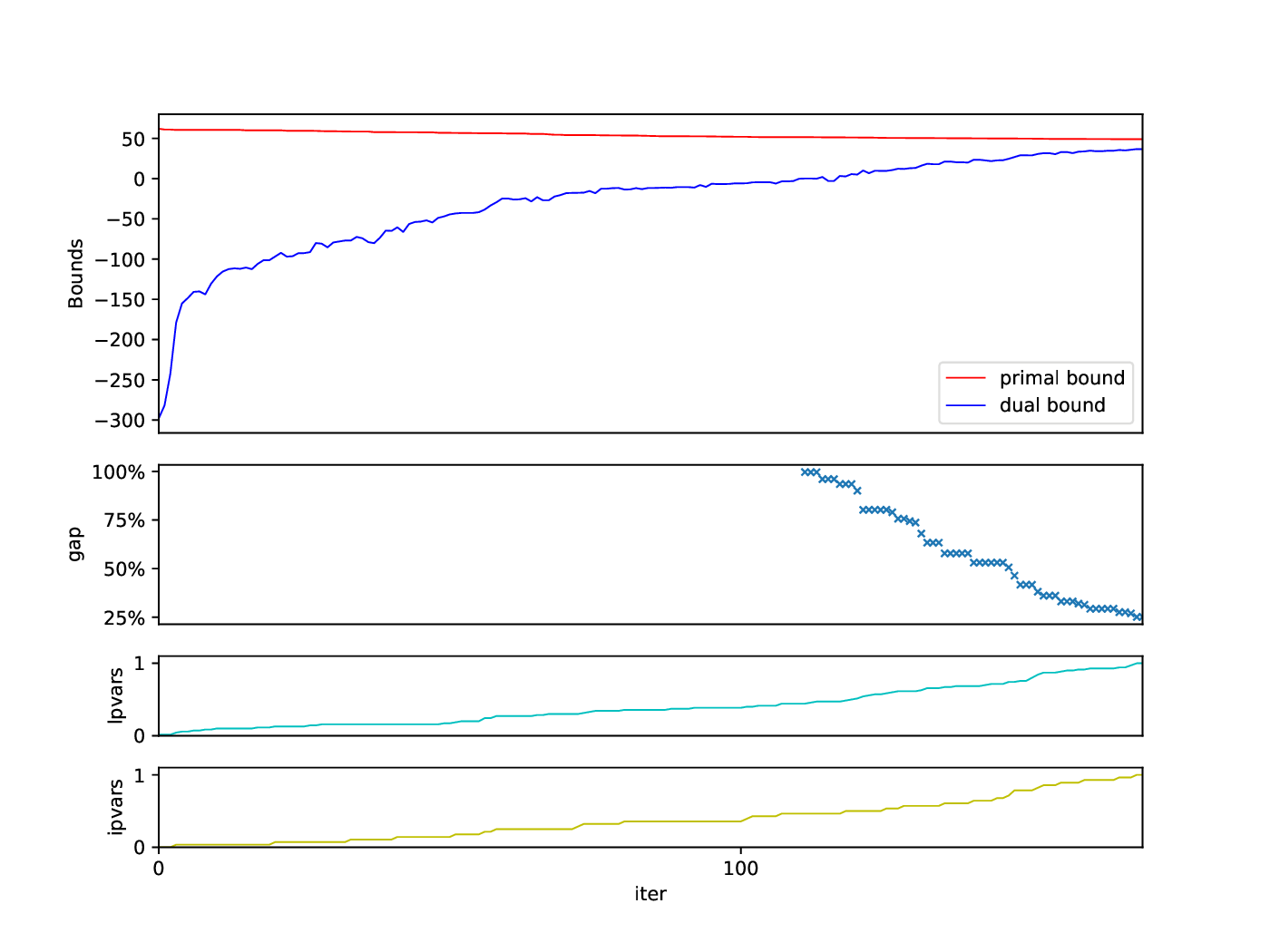
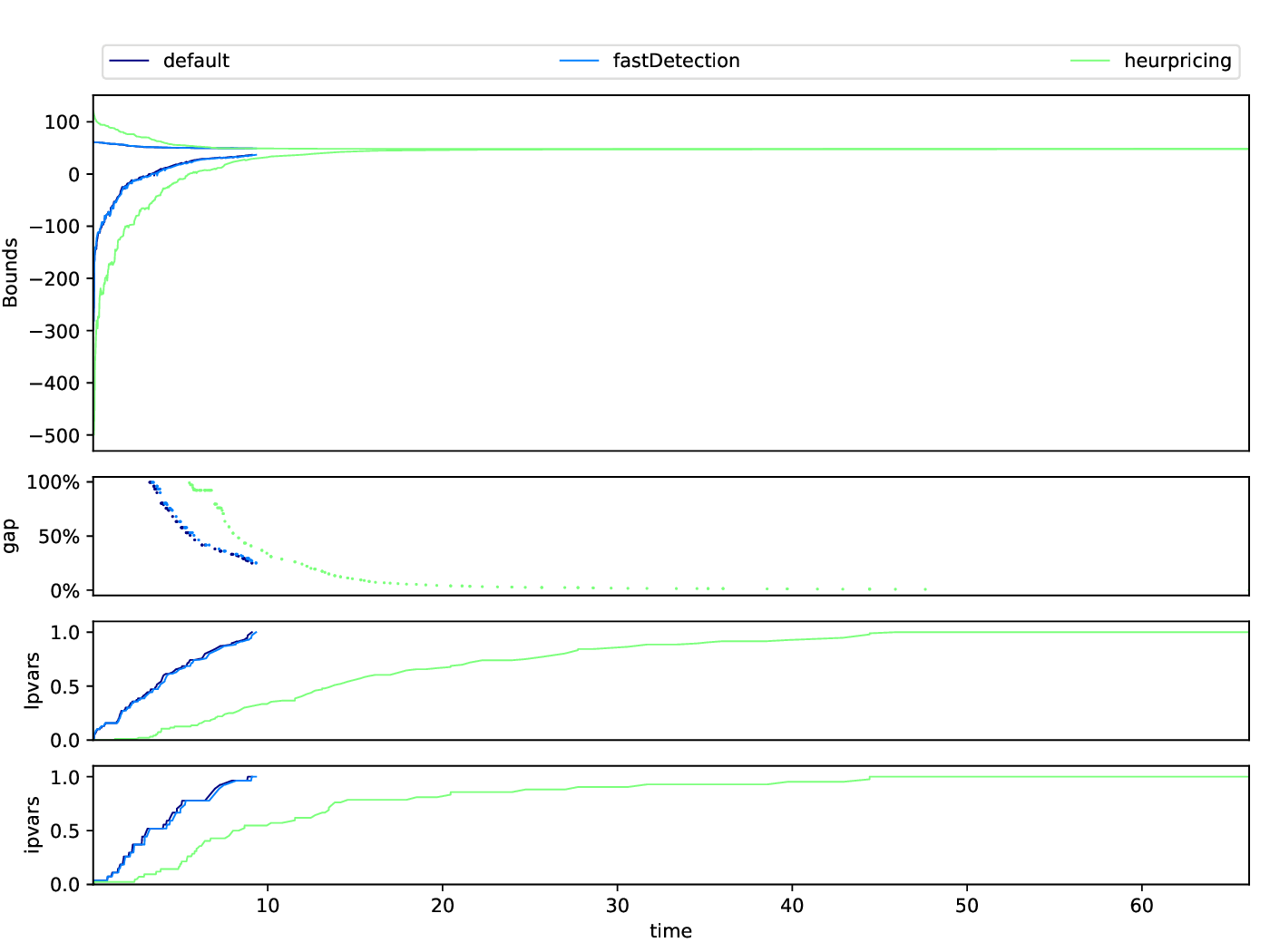
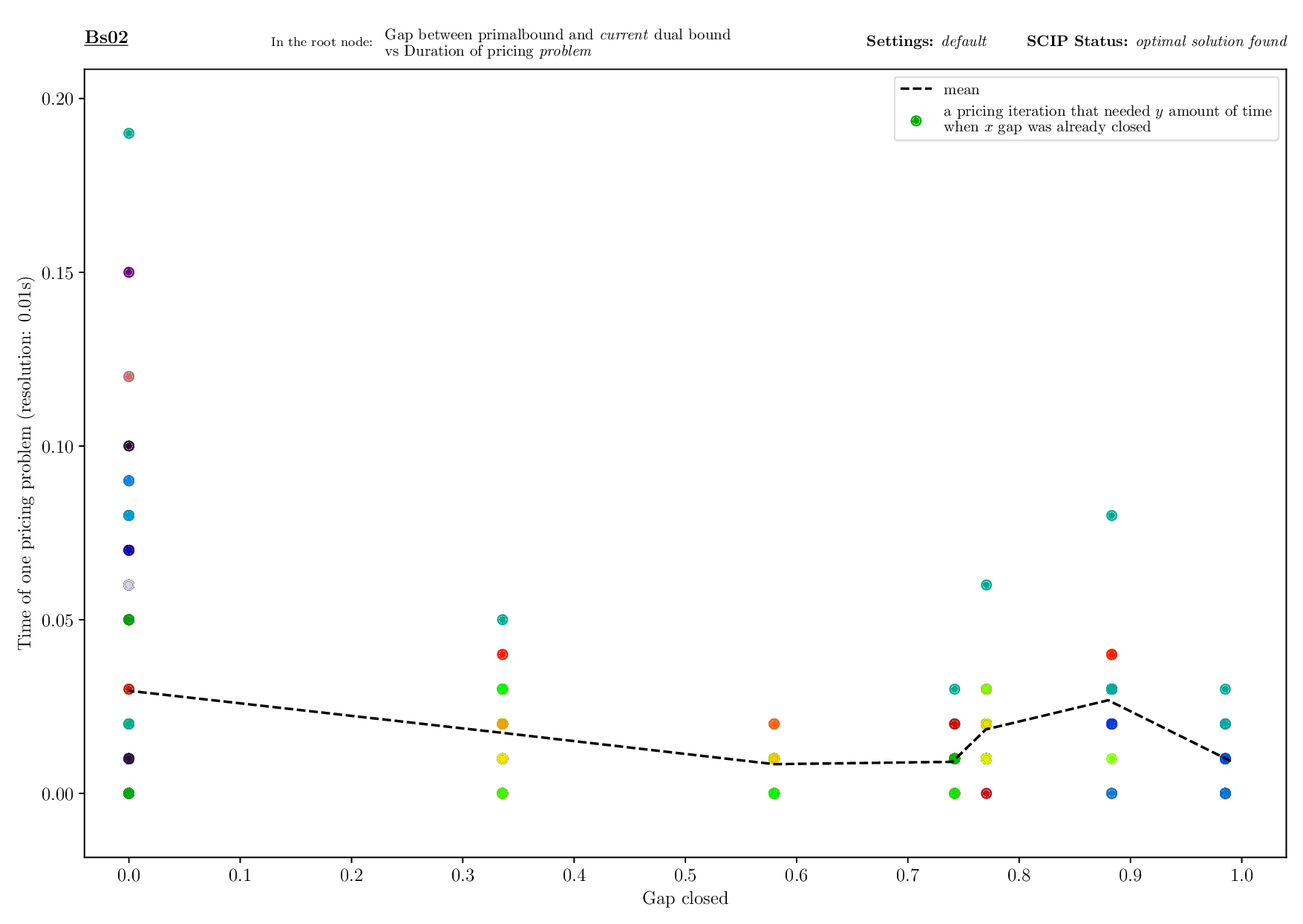
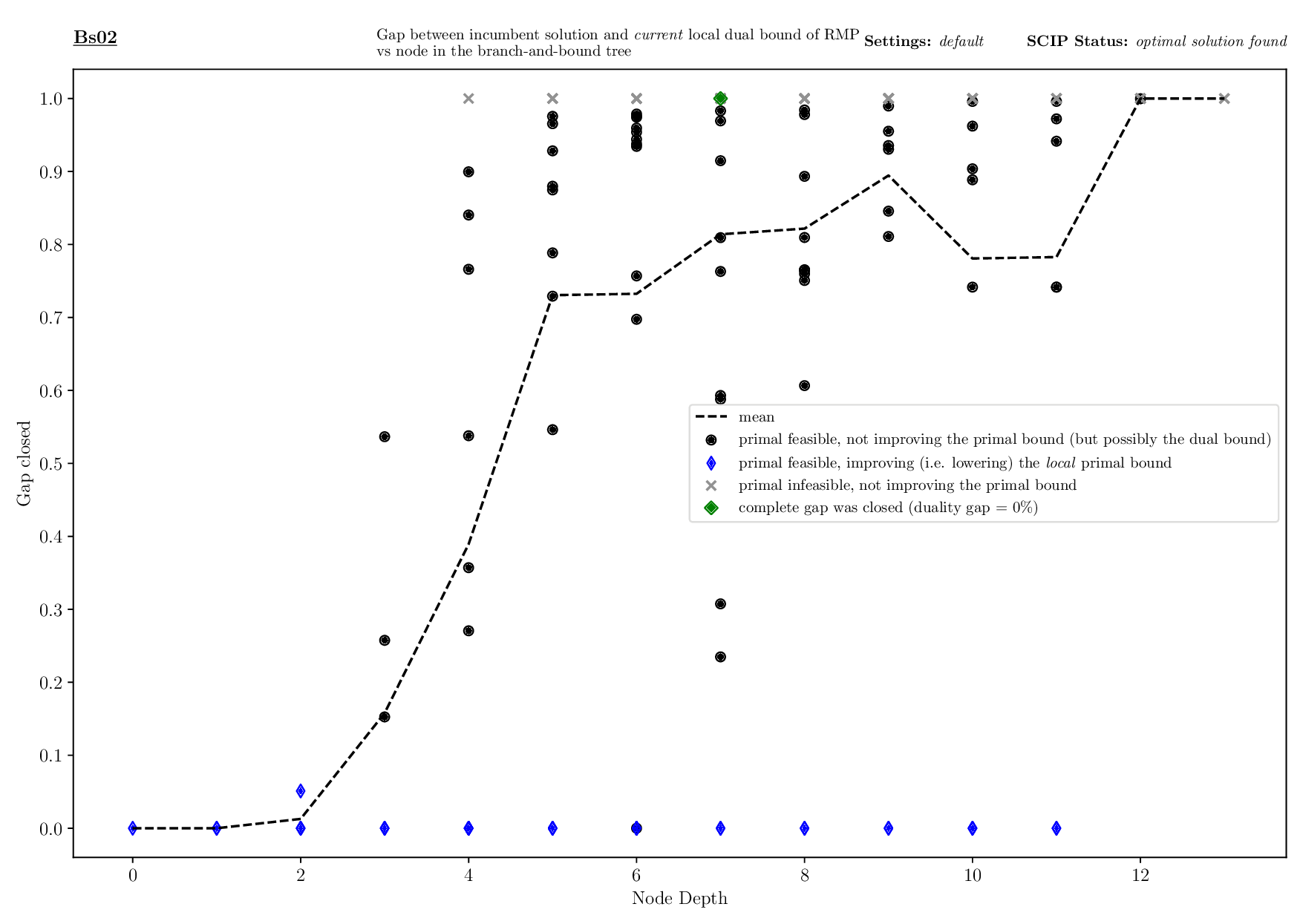
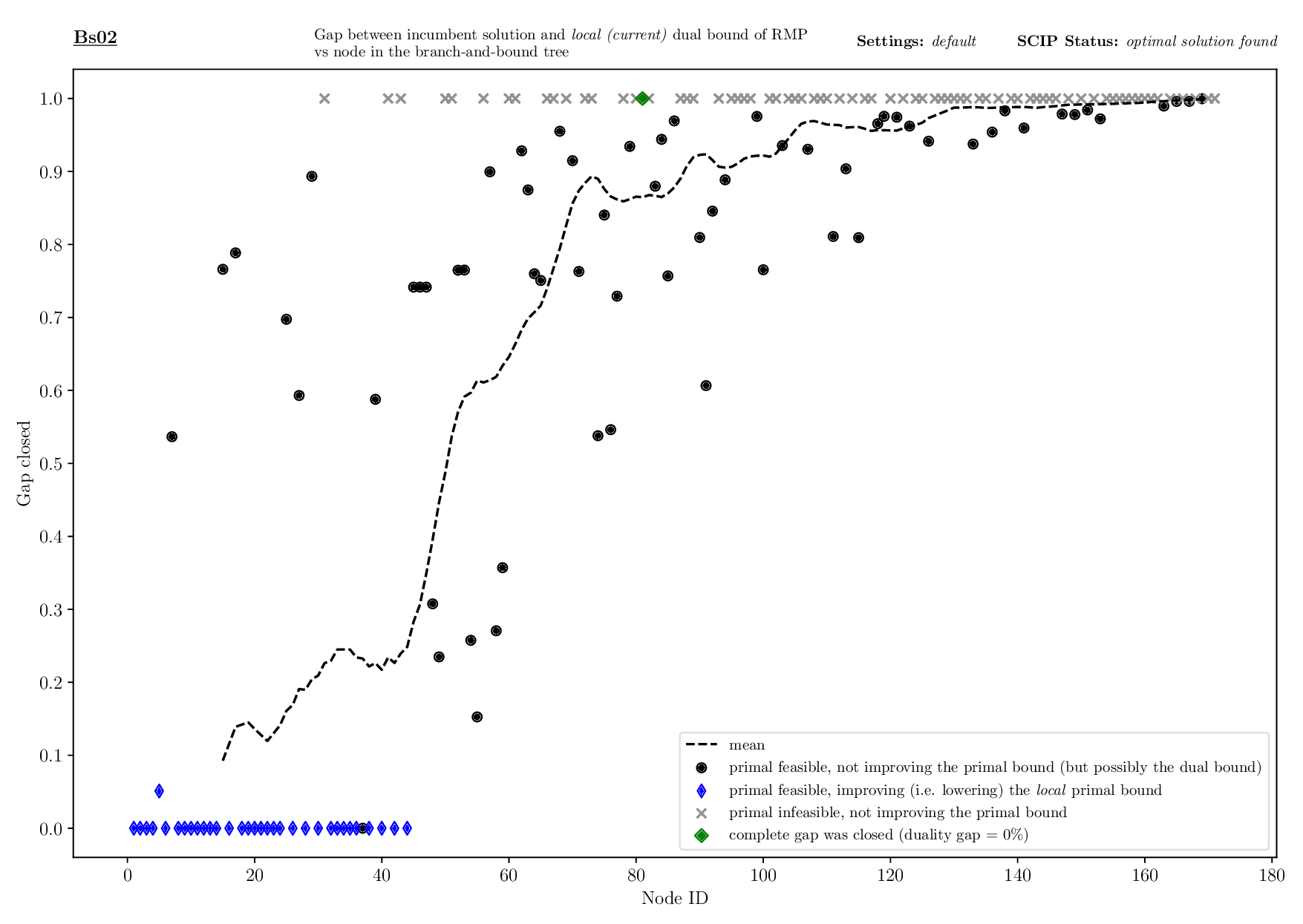
Bounds Plotter
The Bounds Plotter generates a plot showing the development of the primal and dual bound and gap in the root node, as well as the basic variables generated.
Execution
python3 bounds/plotter_bounds.py FILES
with FILES being one or more .out files.
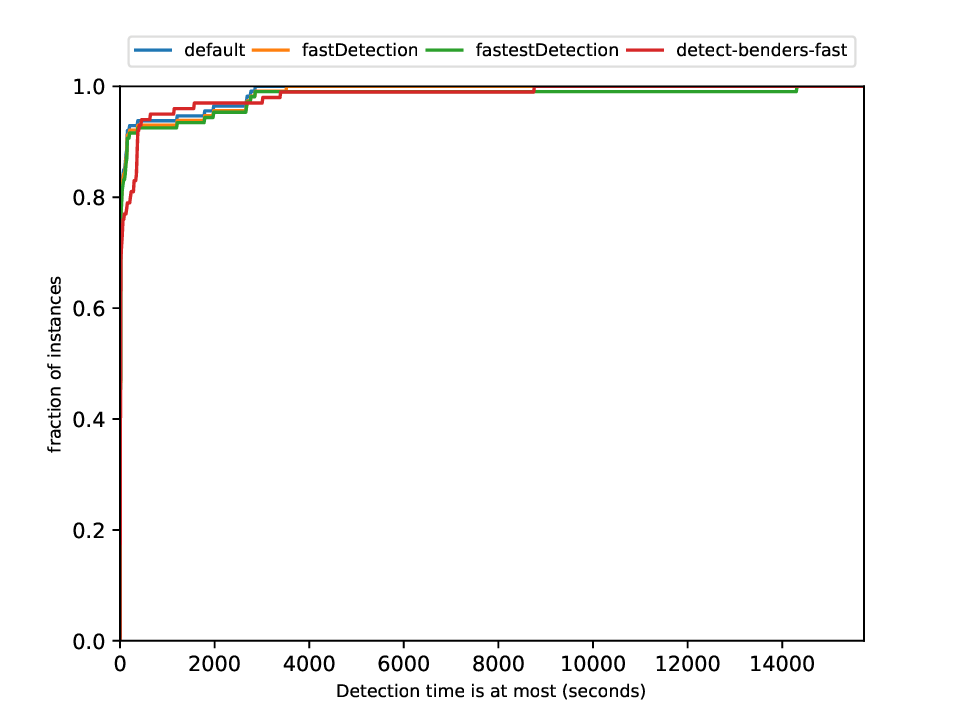
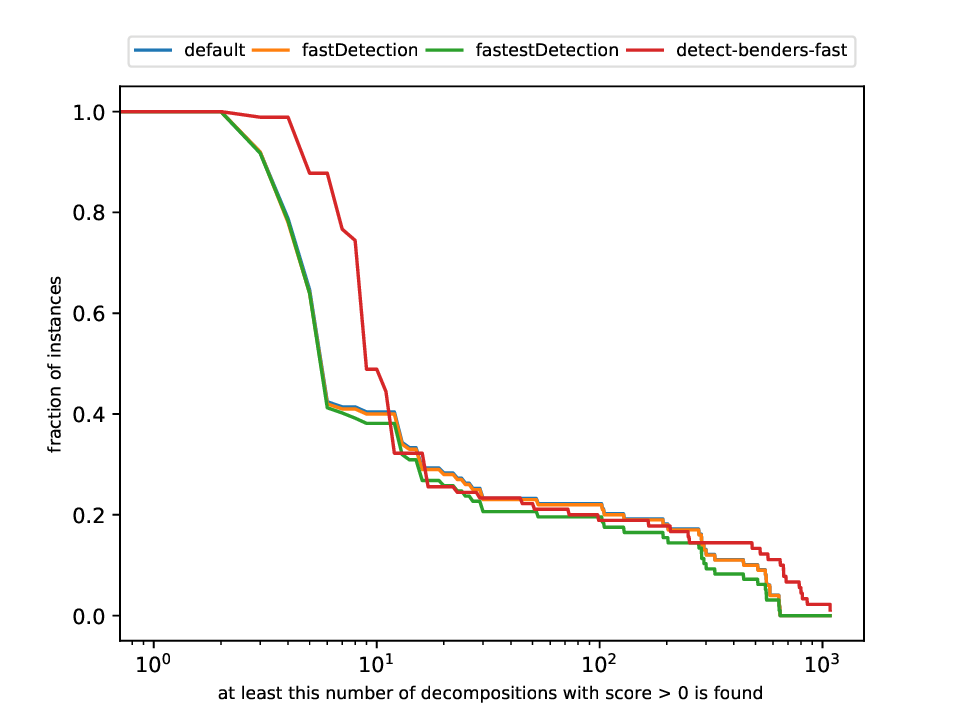
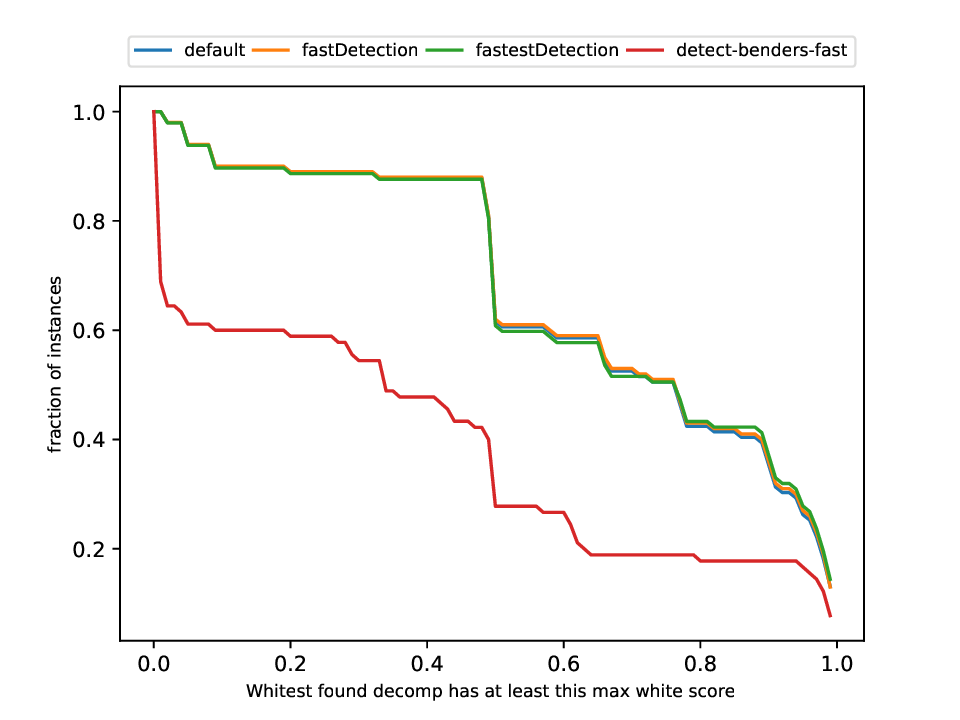
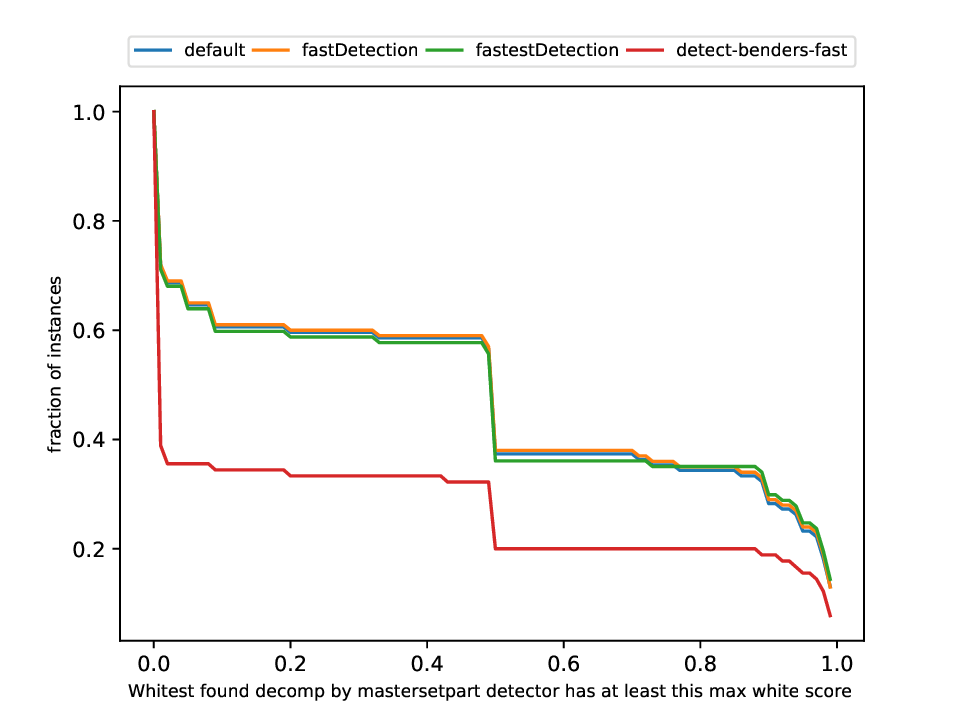
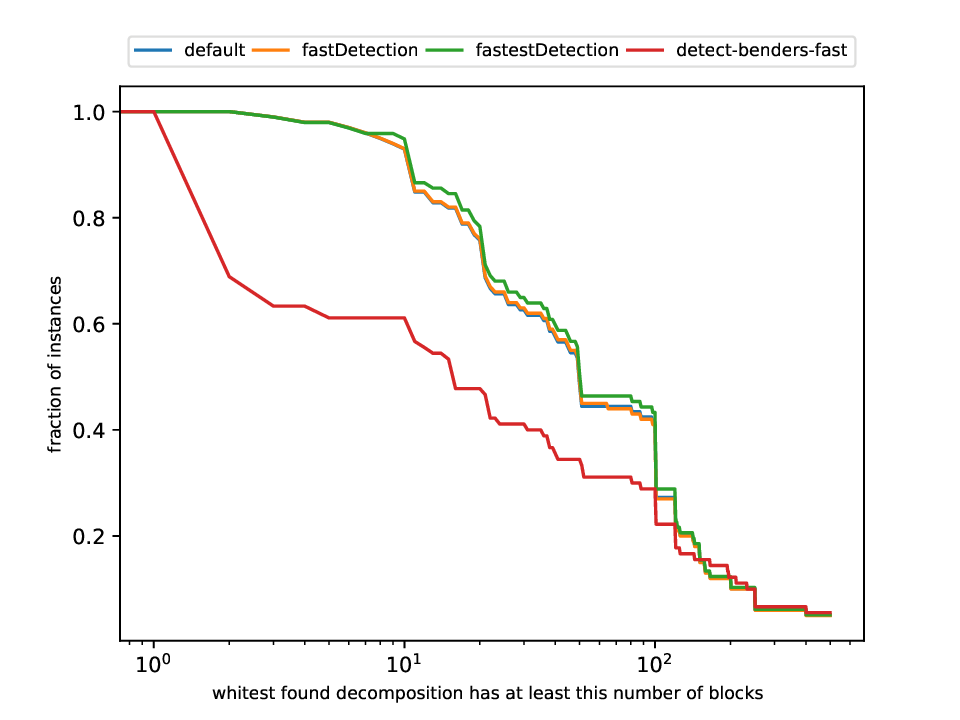
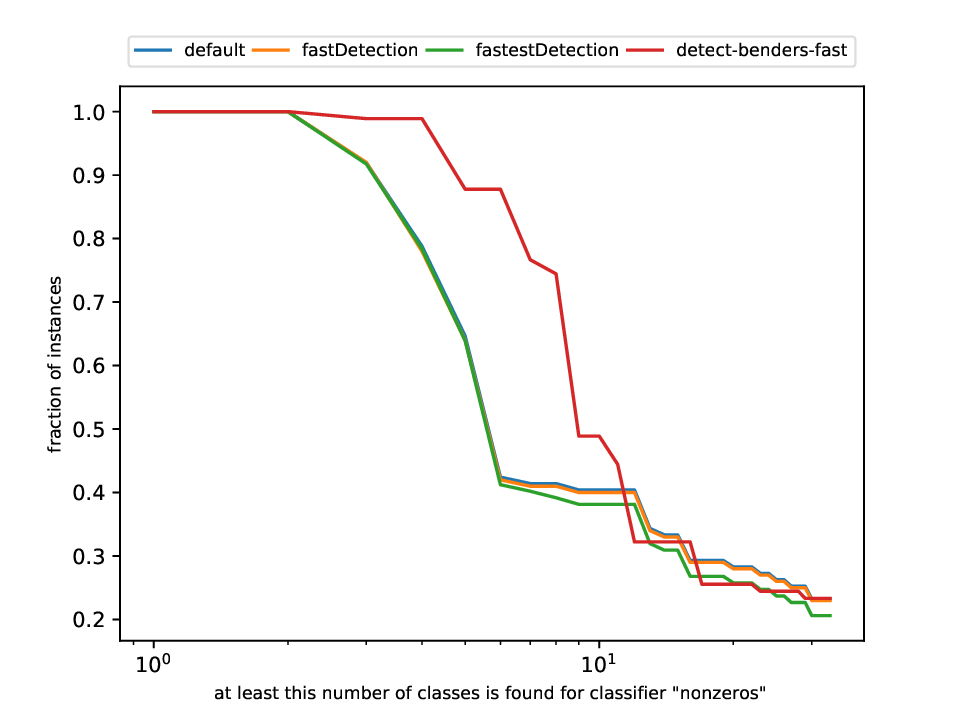
Classification and Detection Plotter
This plotter works on a whole testset and makes plots similar to performance profiles, showing the performance of the classifiers and detectors.
Execution
python3 detection/plotter_detection.py FILES
with FILES being one or more .out files.
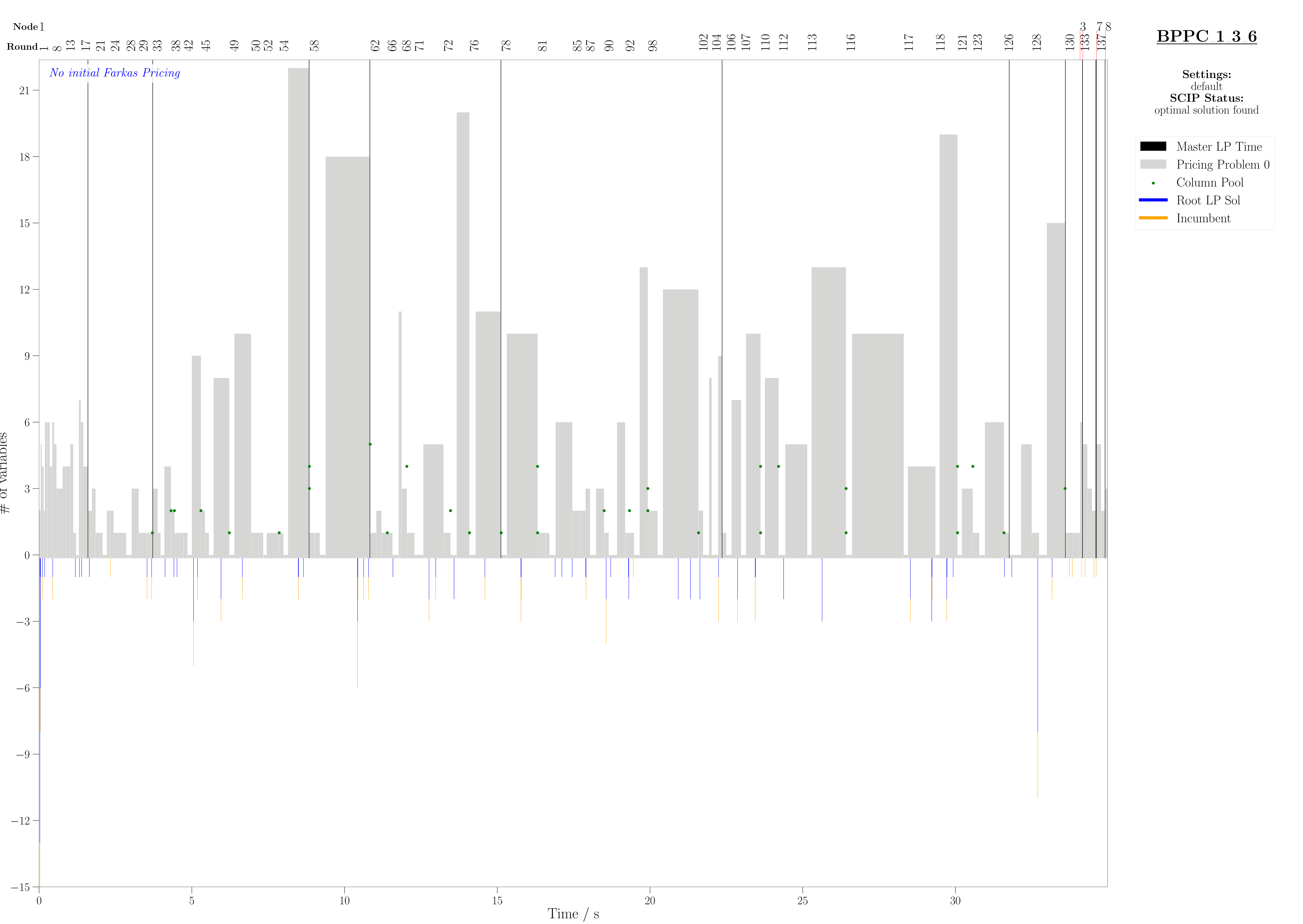
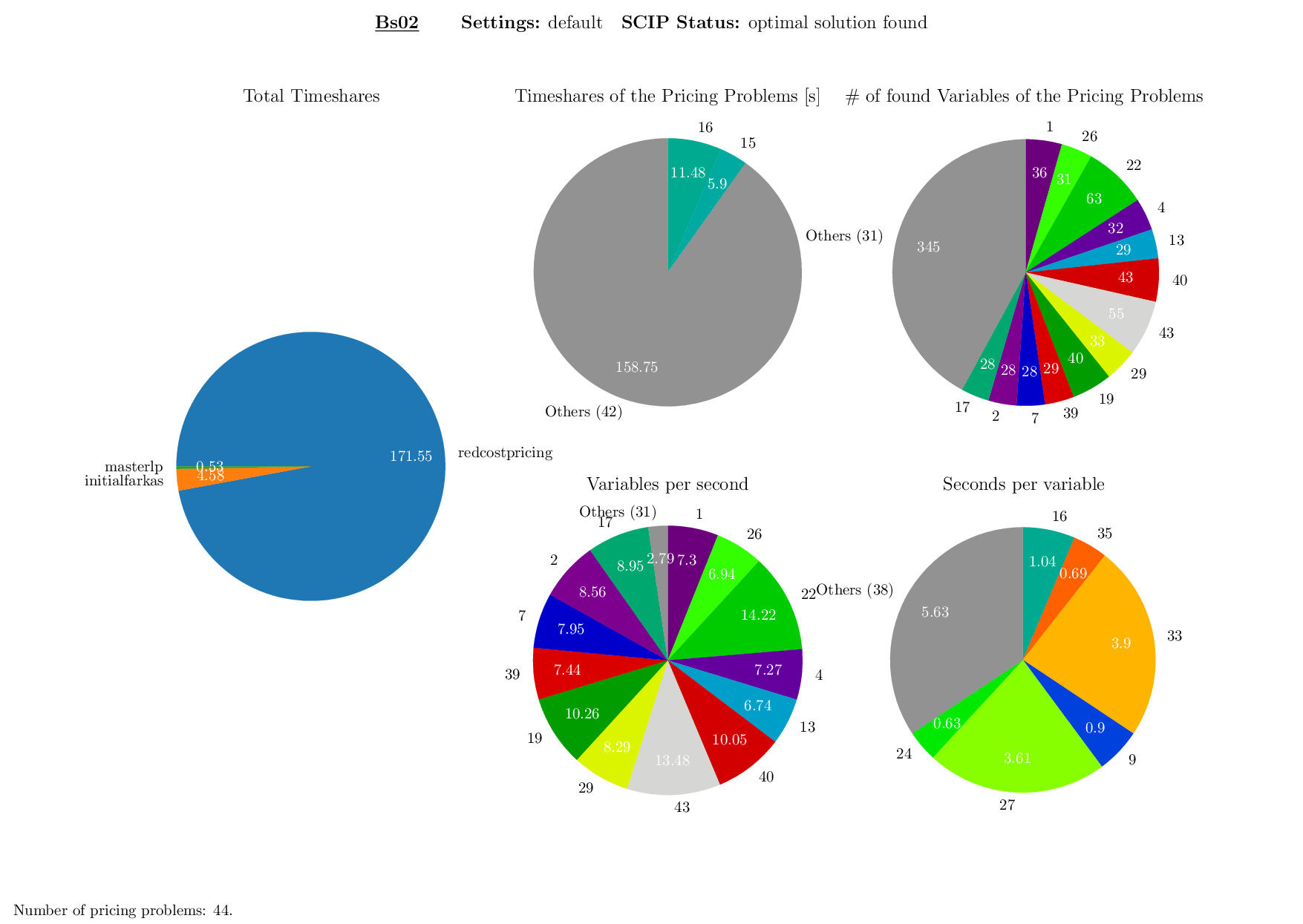
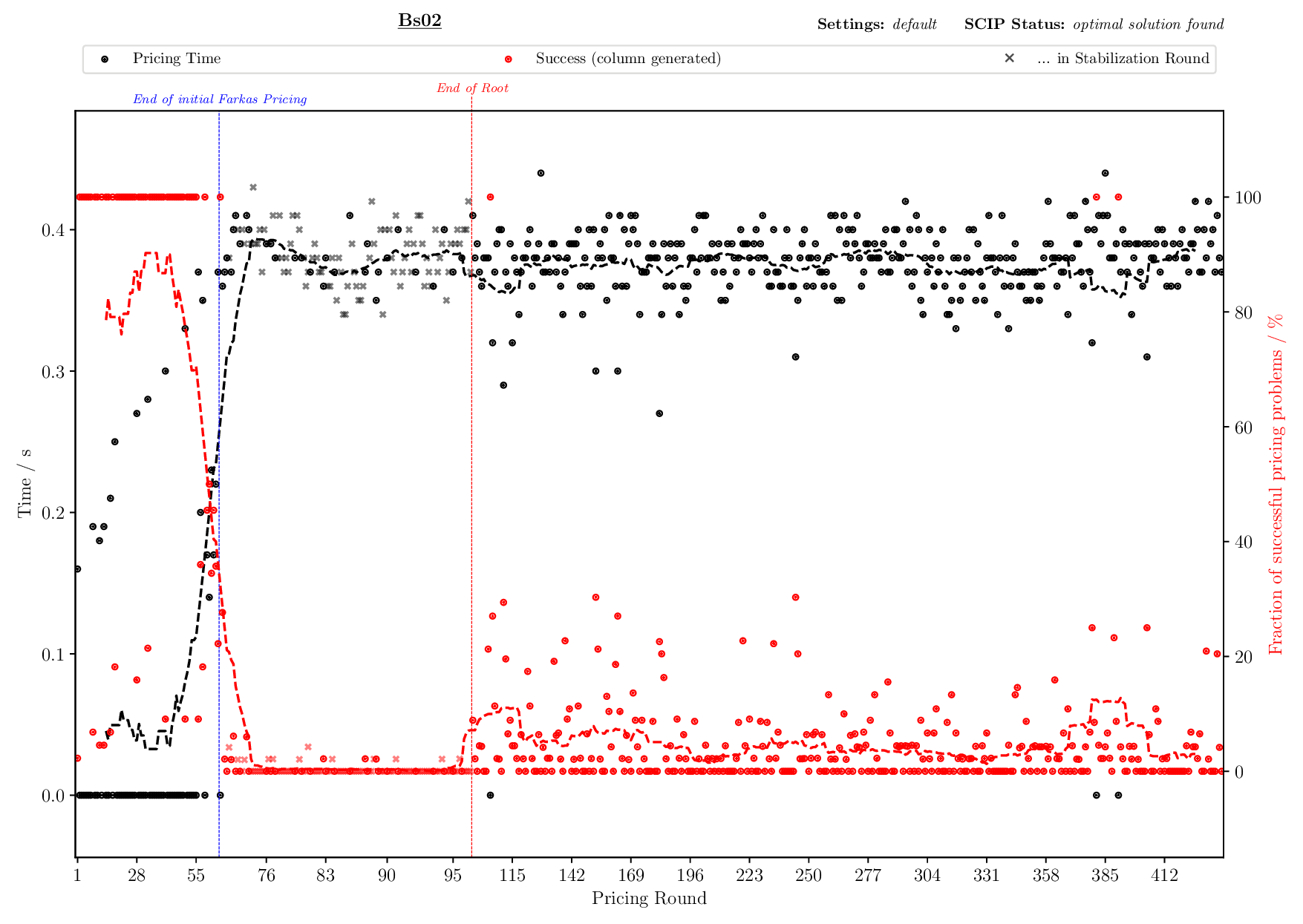
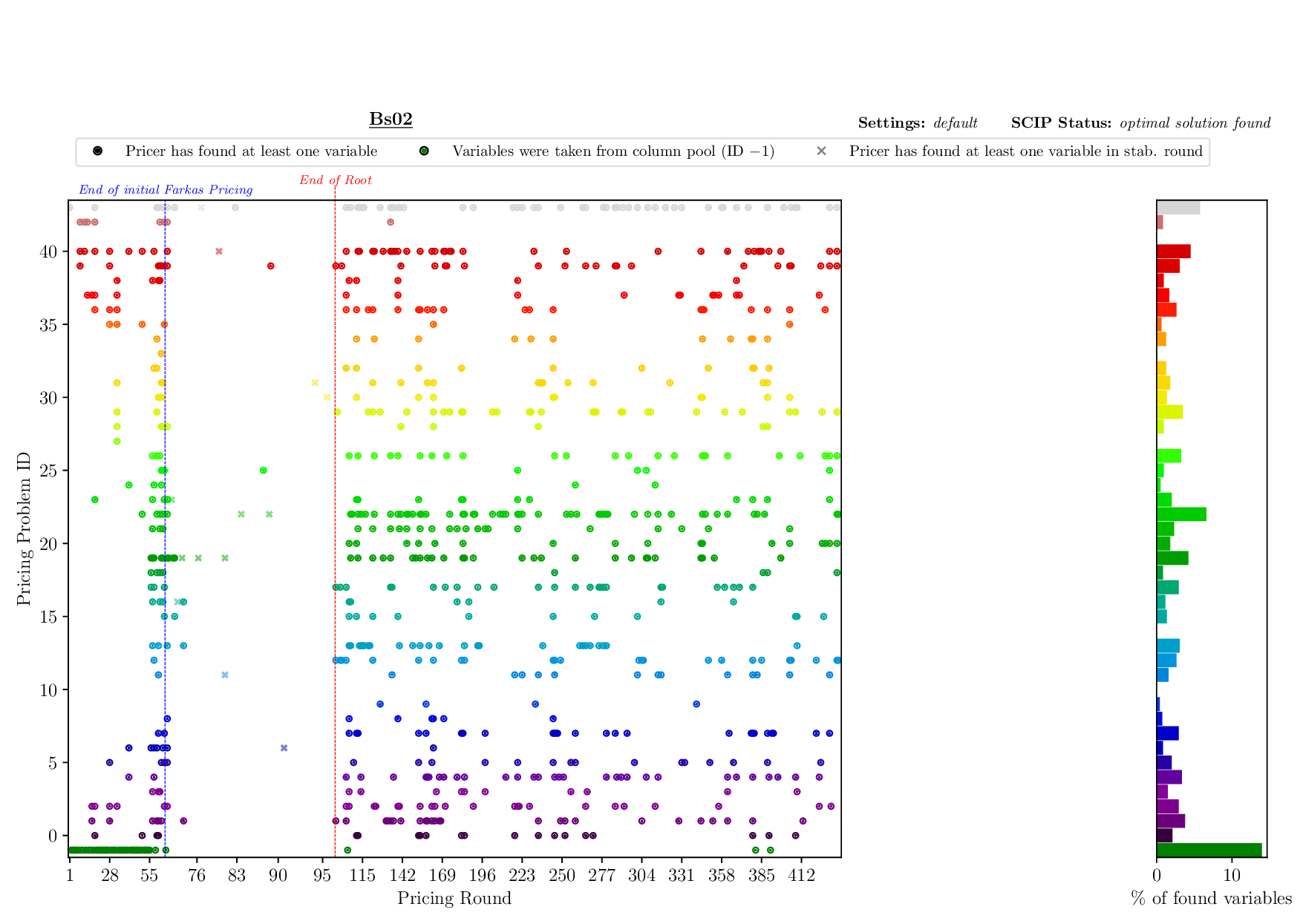
Pricing Plotter
The Pricing Plotter generates 7 different plots illustrating the pricing procedure during a single instance's solving process. When given an outfile with more than one instance, it generates the plots sequentially.
Execution
python3 pricing/plotter_pricing.py FILES --vbcdir VBC
with FILES being one .out file and VBC being the directory where all corresponding .vbc files are (per default: check/results/vbc/)
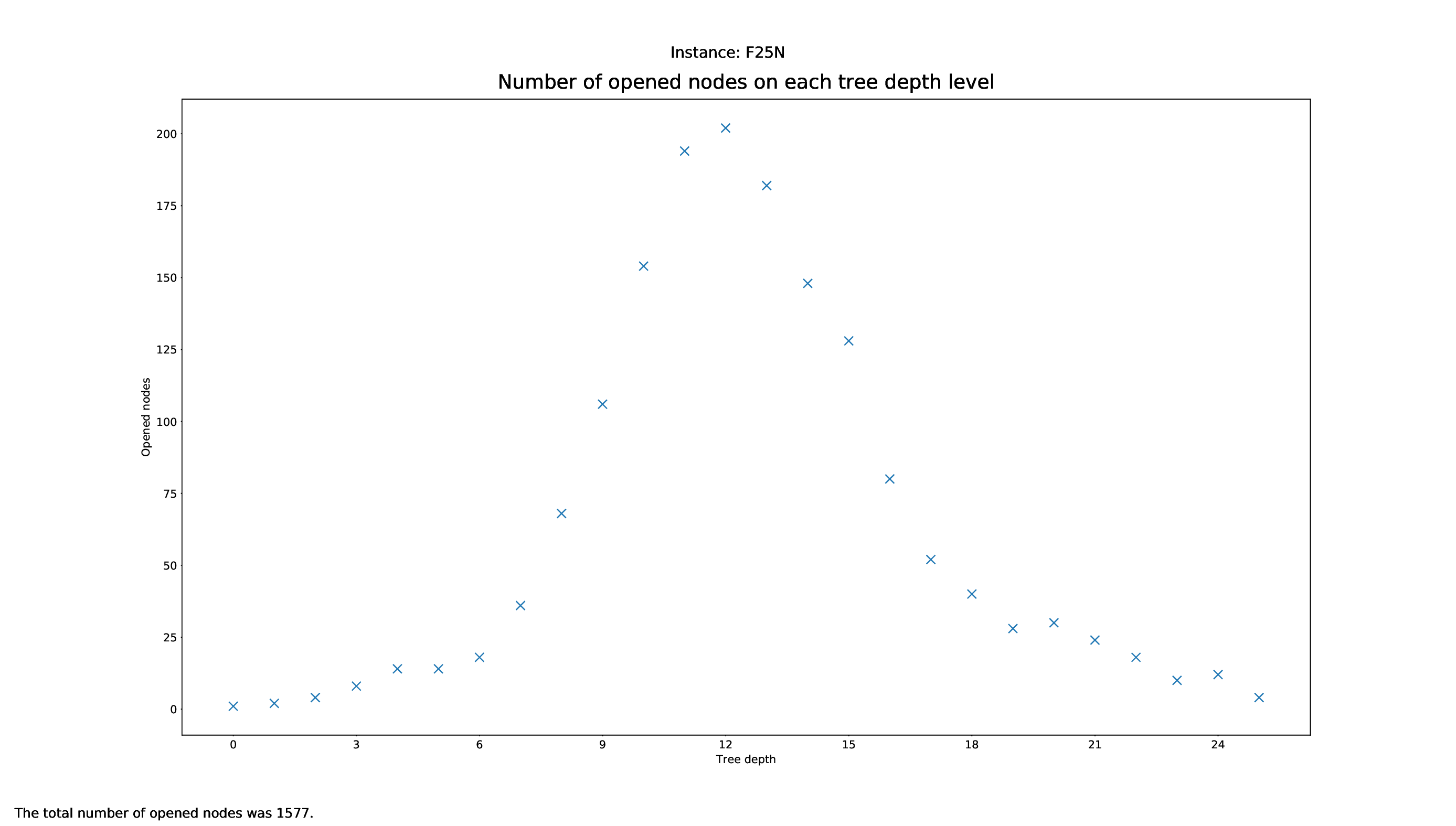
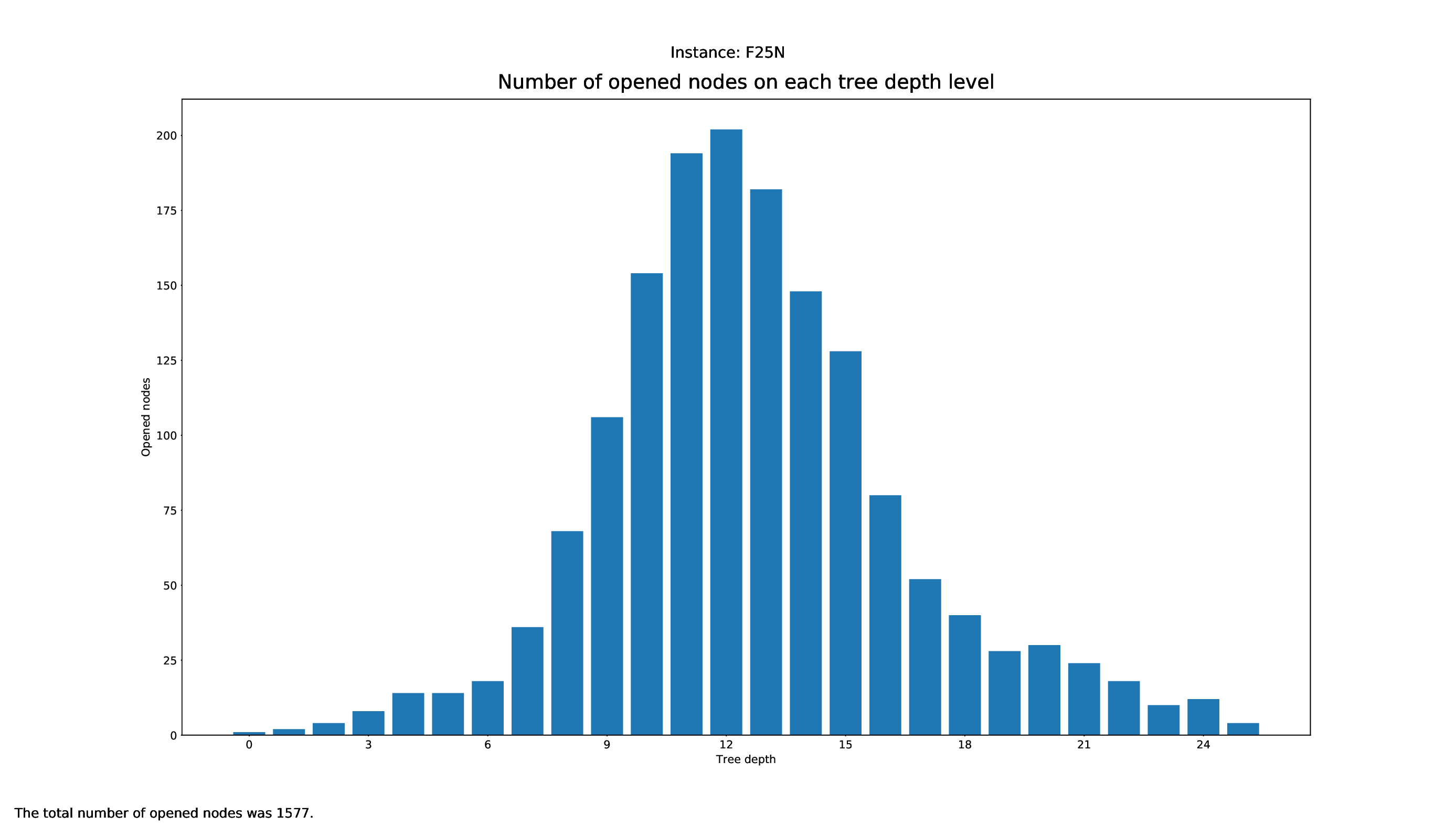
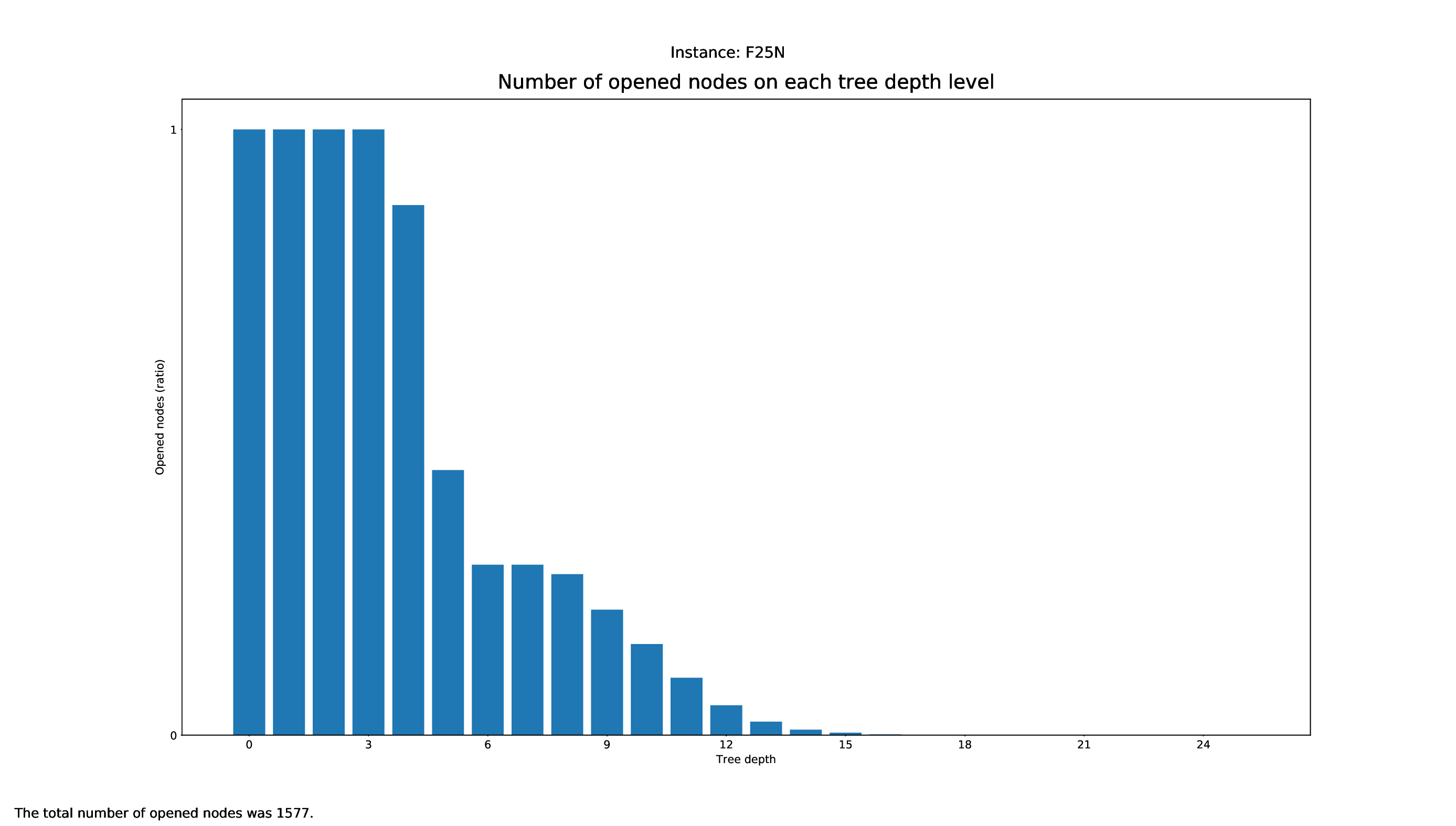
Tree Statistics Plotter
The Tree Statistics Plotter, just like the Pricing Plotter, needs the vbc files to function correctly. It will plot how many nodes were opened on each level.
Execution
python3 tree/plotter_tree.py FILES
with FILES being some .vbc files.
Comparison Table
A (quite raw) comparison of testruns can be done using this script in the general-folder. This script just puts the statistics of all runs that are given as arguments into a .tex-file and prints it as ASCII on the console. Execution
./general/comparison_table.sh run1.res run2.res run3.res ...
with run1, run2, ... being a .res file in the format as shown in 'What files do I get'.
Tree Visualizations
Note: The following guide concerns external software. We do not provide warranty nor support for it. Note: The tree visualization tool (vbctool) does not display aggregated information of the tree's development as the tree statistics plotter does, but shows the tree's development interactively.
In this section, we give a brief guide on how to use a tool to visualize branch-and-cut algorithmics, graphically showing how the tree was built during branching.
Installation
In order to generate pictures of the Branch and Bound tree that GCG used during solving, you can use the vbctool. Since the executable might have issues with the linking of the libraries, it is suggested to download the vbctool source code and additionally the Motif Framework source code, both available on the website. Unzip the Motif Framework source code tarball into the lib/ folder of the vbctool. Before starting with the Build Instructions, install the following packages:
sudo apt-get install libmotif-dev libxext-dev
Then, compile the program (just like explained in the Build Instructions):
cd lib/GFE make cd ../GraphInterface/ make cd ../MotifApp/ make cd ../.. make
Now you can start the program using
./vbctool
Usage
The files you now have to read (File -> Load) are included in the folder check/results/vbc.
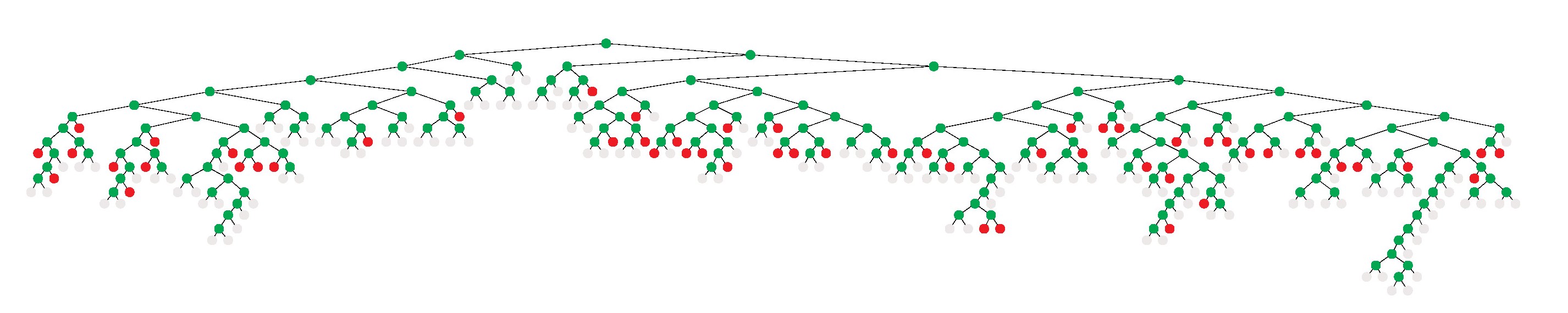
In order to generate the tree, click on Emulation -> Start. Before doing that, you can configure the emulation in Emulation -> Setup, where you can also set the time it will need to generate the tree.
- If left on default values, the tree will generate as fast as it generated in GCG during execution, offering you a good insight into how long GCG was 'stuck' in certain nodes.
- If changed, for example to 1 second, it will just generate the tree all at once and you can then save it.
To save the generated tree, just click on File -> Print and it will save a .ps file.
Test Set Selection
Using existing runtime data, you can filter using the instructions under "General Plotter -> Arguments -> Defining Filters for the Data". For the strIPlib, we can provide a full data set (.out and .pkl format) of runtime data (which is also used in the strIPlib). To then export a test set including the instances that your filter applies to, you can set the flag -ts like that:
python3 general/plot.py check.gcg.out -ts -t "DETECTION TIME" 10 20 "RMP LP TIME"
Note that the test set file export mode is only implemented in the standard plotter, so please call plot.py with your filters and the test set export flag. After executing this command, you will get a test set file filtered.test in the given output directory, with which you can call the make test target as usual. If .dec files were used, they will be included in the exported test set file.
Whenever the test set that you get is too large to use or does not satisfy your requirements, we recommend to check out our diverse test set generation functionality.
Custom Visualizations
When creating custom visualizations, one has to know exactly what data is needed to make the visualization. With these arguments in mind, one can then look if they are already parsed. A list of the currently parsed data is located here. If so, one of the parsers (parser_general.py, parser_bounds.py or parser_detection.py) can be used, or otherwise, for the plotter_pricing.py, the –savepickle argument of the plotter shall be used each time to parse the runtime data and save it to the pickle, to then read it again for the plotter. Then, the plotter_ script can be created which should import the parser that already gets the data needed with a simple import parser_.... Finally, the parser can be used just like in the other plotters.
Troubleshooting
Q: Why don't I get any detection times?
A: You probably did not run the test with a set mode, e.g. MODE=0, so GCG fell back to the readdec mode, reading any .dec files it could find instead of detecting.